10. Scientific and Engineering Libraries¶ Open the notebook in Colab
(C) Copyright Notice: This chapter is part of the book available athttps://pp4e-book.github.io/and copying, distributing, modifying it requires explicit permission from the authors. See the book page for details:https://pp4e-book.github.io/
In this chapter, we will cover several libraries that are very functional in scientific & engineering-related computing problems. In order to keep our focus on the practical usages of these libraries and considering that this is an introductory textbook, the coverage in this chapter is in not intended to be comprehensive.
10.1. Numerical Computing with NumPy¶
NumPy can be considered as a library for working with vectors and matrices. NumPy calls vectors and matrices as arrays.
Installation Notes To be able to use the NumPy library, you will need to download it from
numpy.org and install it on your computer. If
you are using a Python package manager (e.g. pip), you can install it
directly using: |
10.1.1. Arrays and Their Basic Properties¶
Let us consider a simple vector and a matrix:
and
Let us see how we can represent and work with these two arrays in NumPy:
>>> import numpy as np # Import the NumPy library
>>> array1 = np.array([1, 2, 3])
>>> array2 = np.array([[1, 2, 3], [4, 5, 6]])
>>> type(array1)
<class 'numpy.ndarray'>
>>> type(array2)
<class 'numpy.ndarray'>
>>> array1
array([1, 2, 3])
>>> array2
array([[1, 2, 3],
[4, 5, 6]])
>>> print(array2)
[[1 2 3]
[4 5 6]]
We see from this example that we can pass lists of number as arguments
to np.array function which creates a NumPy array for us. If the
argument is a nested list, each element of the list is used as a row of
a 2D array.
Arrays can contain any data type as elements; however, we will limit ourselves to numbers (integers and real numbers) in this chapter.
Shapes, Dimensions and Number of Elements of Arrays. The first thing
we can do with a NumPy array is check its shape. For our example
array1 and array2, we can do so as follows:
>>> array1.shape
(3,)
>>> array2.shape
(2,3)
where we can see that array1 is a one-dimensional array with
\(3\) elements and array2 is a \(2\times 3\) array (a 2D
matrix).
For a 2D array, a shape value (R, C) denotes the number of rows
first (R), which is sometimes also called the first dimension, and
then the number of columns (C), which is the second dimension. For
\(n\)D arrays with \(n>2\), the meaning of the shape values is
the same except that there are \(n\) values in the shape.
In NumPy, we can easily change the shape of an array without losing content:
>>> array1.reshape((3,1))
array([[1],
[2],
[3]])
>>> array2.reshape((1,6))
array([[1, 2, 3, 4, 5, 6]])
For many applications, we will need to access the number of dimensions
of an array. For this purpose, we can use .ndim value:
>>> array1.ndim
1
>>> array2.ndim
2
The number of elements in an array is another important value that we
are frequently interested in. To access that, we can use the .size
value:
>>> array1.size
3
>>> array2.size
6
Accessing elements in arrays. NumPy allows the same indexing mechanisms that you can use with Python’s native container data types. For NumPy, let us look at some examples:
>>> array1[-1]
3
>>> array2[1][2]
6
>>> array2[-1]
array([4, 5, 6])
Creating arrays. We have already seen that we can create arrays
using the np.array() function. However, there are other ways for
creating arrays conforming to predefined specifications. We can for
example create arrays filled with zeros or ones (note that the argument
is a tuple describing the shape of the matrix):
>>> np.zeros((3, 4))
array([[0., 0., 0., 0.],
[0., 0., 0., 0.],
[0., 0., 0., 0.]])
>>> np.ones((2,6))
array([[1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1.]])
Alternatively, we can create an array filled with a range of values
using np.arange() function:
>>> np.arange(1,10) # 1: starting value. 10: ending value (excluded)
array([1, 2, 3, 4, 5, 6, 7, 8, 9])
>>> np.arange(1,10,2)
array([1, 3, 5, 7, 9])
>>> np.arange(1,10).reshape((3,3))
array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
10.1.2. Working with Arrays¶
The previous section covered how we can access elements in an array and properties of an array. Now let us see how the different types of operations we can do with arrays.
Arithmetic, Relational and Membership Operations with Arrays
The arithmetic operations (+, -, *, /, **), the
relational operations (==, <, <=, >, >=) and the
membership operations (in, not in) that we can apply on numbers
and other data types in Python can be applied on arrays with NumPy.
These operations are performed elementwise. This means that the arrays
that are provided as operands to a binary arithmetic operator need to
have the same shape.
Let us see some examples for arithmetic operations:
>>> A = np.arange(4).reshape((2,2))
>>> A
array([[0, 1],
[2, 3]])
>>> B = np.arange(4, 8).reshape((2,2))
>>> B
array([[4, 5],
[6, 7]])
>>> print(B-A)
[[4 4]
[4 4]]
>>> print(B+A)
[[ 4 6]
[ 8 10]]
>>> A
array([[0, 1],
[2, 3]])
>>> B
array([[4, 5],
[6, 7]])
Note here that these operations create a new array whose elements are
the results of applying the operation. Therefore, the original arrays
are not modified. If you are interested in in-place operations that
modify an existing array during the operation, you can use combined
statements such as +=, -=, *=.
Relational and membership operations are also applied elementwise and we can easily anticipate the outcomes of such operations, e.g. as follows:
>>> A < B
array([[ True, True],
[ True, True]])
>>> B > A
array([[ True, True],
[ True, True]])
>>> A > B
array([[False, False],
[False, False]])
>>> 4 in B
True
>>> 10 in B
False
Useful Functions
NumPy arrays provide several useful functions already provided. These
include: - Standard mathematical function such as exponent, sin, cos,
square-root: np.exp(<array>), np.sin(<array>),
np.cos(<array>), np.sqrt(<array>). - Minimum and maximum:
<array object>.min() and <array object>.max(). - Summation, mean
and standard deviation: <array object>.sum(),
<array object>.mean() and <array object>.std().
Note that minimum, maximum, summation, mean and standard deviation can
be applied on the whole array as well as along a pre-specified dimension
(specified with an axis parameter). Let us see some examples to
clarify this important aspect:
>>> A
array([[0, 1],
[2, 3]])
>>> A.sum()
6
>>> A.sum(axis=0)
array([2, 4])
>>> A.sum(axis=1)
array([1, 5])
>>> A.sum(axis=2)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/usr/local/lib/python3.7/site-packages/numpy/core/_methods.py", line 47, in _sum
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
numpy.AxisError: axis 2 is out of bounds for array of dimension 2
where we see that axes start being numbered from zero.
Splitting and Combining Arrays
For many problems, we will need to split an array into multiple arrays
or combine multiple arrays into one. For splitting arrays, we can use
functions such as np.hsplit (for horizontal split), np.vsplit
(for vertical split) and np.array_split (for more general split
operations). Below is an example for hsplit and vsplit:
>>> L = np.arange(16).reshape(4,4)
>>> L
array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11],
[12, 13, 14, 15]])
>>> np.hsplit(L,2) # Divide L into 2 arrays along the horizontal axis
[array([[ 0, 1],
[ 4, 5],
[ 8, 9],
[12, 13]]), array([[ 2, 3],
[ 6, 7],
[10, 11],
[14, 15]])]
>>> np.vsplit(L,2)
[array([[0, 1, 2, 3],
[4, 5, 6, 7]]), array([[ 8, 9, 10, 11],
[12, 13, 14, 15]])]
Note that the resultant arrays are provided as a Python list. Note
also that hsplit and vsplit functions work on even sizes
(i.e. they can split into equally sized arrays) – for general split
operations, you can use array_split.
For combining multiple arrays, we can use np.hstack (for horizontal
stacking), np.vstack (for vertical stacking) and np.stack (for
more general stacking operations). Below is an example (for the A
and B arrays that we have created before):
>>> A
array([[0, 1],
[2, 3]])
>>> B
array([[4, 5],
[6, 7]])
>>> np.hstack((A, B))
array([[0, 1, 4, 5],
[2, 3, 6, 7]])
>>> np.vstack((A, B))
array([[0, 1],
[2, 3],
[4, 5],
[6, 7]])
Iterations with Arrays.
NumPy library already provides for us many functionalities that we might need while working with arrays. However, in many circumstances those will not be necessary and we will need to be able to iterate over the elements of arrays and perform custom algorithmic steps.
Luckily, iteration with arrays is very similar to how we would perform iterations with other container data types in Python. Let us see some examples:
>>> L
array([[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11],
[12, 13, 14, 15]])
>>> for r in L:
... print("row: ", r)
...
row: [0 1 2 3]
row: [4 5 6 7]
row: [ 8 9 10 11]
row: [12 13 14 15]
where we see that iteration over a multi-dimensional array iterates over the first dimension. To iterate over each element, we have at least two options:
>>> for element in L.flat:
... print(element)
...
0
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
>>> for r in L:
... for element in r:
... print(element)
...
0
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
Now let us implement one of the operations we have seen above in Python from scratch as an exercise:
import numpy as np
def horizontal_stack(A,B):
"""A function that combines two 2D arrays A and B.
A and B need to have the same height.
"""
# Let us get dimensions first and check whether A and B
# are compatible for stacking.
(H_A, W_A) = A.shape
(H_B, W_B) = B.shape
if H_A != H_B:
print("Arguments A and B have incompatible heights!")
return None
# Let us create an empty `result` array:
H_result = H_A #or H_B
W_result = W_A + W_B
result = np.zeros((H_result, W_result))
# Now let us iterate over each position in A and B and place
# their elements into the corresponding positions
for i in range(H_A):
for j in range(W_A):
result[i][j] = A[i][j]
for j in range(W_B):
result[i][j+W_A] = B[i][j]
return result
# Let us test our code:
M = np.random.randn(2,2) # Create a random 3x4 array
N = np.random.randn(2,3) # Create a random 3x6 array
print("Random array M is:\n", M)
print("Random array N is:\n", N)
print("Horizontal stacking of M and N yields:\n", horizontal_stack(M, N))
Random array M is:
[[ 0.39521366 1.41939618]
[-0.13770429 1.19319949]]
Random array N is:
[[-1.61066212e-01 8.15354855e-01 7.19234023e-01]
[ 2.65985989e-04 -2.89111500e-01 4.52781848e-02]]
Horizontal stacking of M and N yields:
[[ 3.95213656e-01 1.41939618e+00 -1.61066212e-01 8.15354855e-01
7.19234023e-01]
[-1.37704293e-01 1.19319949e+00 2.65985989e-04 -2.89111500e-01
4.52781848e-02]]
10.1.3. Linear Algebra with NumPy¶
Now let us give a flavour of the Linear Algebra operations provided by
NumPy. Note that some of these operations is provided in the module
numpy.linalg.
Transpose.
The transpose operation flips a matrix along the diagonal. For an example matrix \(A\),
its transpose is the following:
In NumPy, the transpose of a matrix can be easily accessed by accessing
its .T member variable or by calling its .transpose() member
function:
>>> A
array([[1, 2],
[3, 4]])
>>> A.T
array([[1, 3],
[2, 4]])
>>> A
array([[1, 2],
[3, 4]])
Note that .T is simply a member variable of the array object and is
not defined as an operation. Try finding the transpose of A with
A.transpose() and see that you obtain the expected result. Note also
that the transpose operation does not change the original array.
Inverse.
The inverse of an \(n\times n\) matrix \(A\) is an \(n\times n\) matrix \(A^{-1}\) that yields an \(n\times n\) identity matrix \(I\) when multiplied:
In NumPy, we can use np.linalg.inv(<array>) to find the inverse of a
square array. Here is an example:
>>> A
array([[1, 2],
[3, 4]])
>>> A_inv = np.linalg.inv(A)
>>> A_inv
array([[-2. , 1. ],
[ 1.5, -0.5]])
Let us check whether the inverse was correctly calculated:
>>> np.matmul(A, A_inv)
array([[1.0000000e+00, 0.0000000e+00],
[8.8817842e-16, 1.0000000e+00]])
which is the identity matrix (\(I\)) except for some rounding error
at index \((1,0)\) owing to floating point approximation. However,
as we have seen in Chapter 2, while writing our programs, we should
compare numbers such that 8.8817842e-16 can be considered to be
equal to zero.
Determinant, norm, rank, condition number, trace
While working with matrices, we often require the following properties which can be easily calculated. For the sake of brevity and to keep the focus, we will omit their explanations:
Matrix Property |
How to Calculate with NumPy |
|---|---|
Determinant (\(|A|\) or \(det(A)\)) |
|
Norm (\(||A||\)) |
|
Rank (\(rank(A)\)) |
|
Condition number (\(\kappa(A)\)) |
|
Trace (\(tr(A)\)) |
|
Dot Product, Inner Product, Outer Product, Matrix Multiplication
The product of two matrices can be calculated in different ways:
np.dot(a, b): For vectors (1D arrays), this is equivalent to the dot product (i.e. \(\sum_i \mathbf{a}_i \times \mathbf{b}_i\) for two vectors \(\mathbf{a}\) and \(\mathbf{b}\)). For 2D arrays and arrays with more dimensions, the result is matrix multiplication.np.inner(a,b): For vectors, this is dot product (likenp.dot(a,b)). For higher-dimensional arrays, the result is a sum product over the last axes. Consider the following example for clarification on this:
>>> A
array([[0, 1],
[2, 3]])
>>> B
array([[4, 5],
[6, 7]])
>>> np.inner(A,B)
array([[ 5, 7],
[23, 33]])
The last axes for both A and B are the horizontal axes.
Therefore, each first-axis element of A (i.e. [0, 1] and [2,3]) is
multiplied with each first-axis element of B ([4, 5] and [6, 7]).
np.outer(a,b): Outer product is defined on vectors such thatresult[i,j] = a[i] * b[j].np.matmul(a,b): Matrix multiplication of matricesaandb. The result is simply calculated as \(\textrm{result}_{ij} = \sum_k \mathbf{a}_{ik} \mathbf{b}_{kj}\), as also illustrated in Fig. 10.1.1.
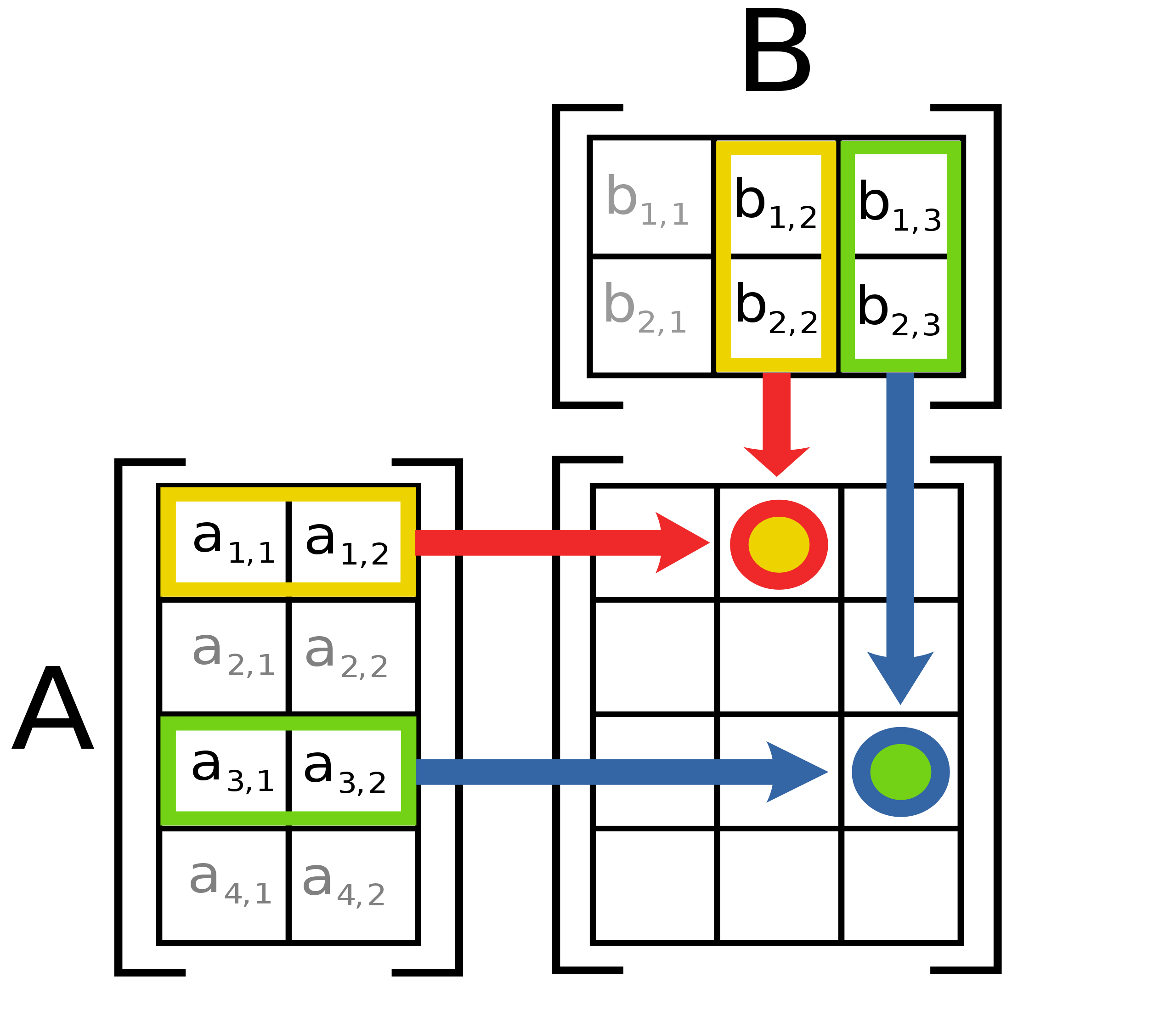
Fig. 10.1.1 Illustration of matrix multiplication of two matrices A and B. (Figure source: Wikipedia)¶
Eigenvectors and Eigenvalues
Explaining eigenvectors and eigenvalues is beyond the scope of the book. However, we can briefly remind that eigenvectors and eigenvalues are the variables that satisfy the following for a matrix \(A\) which can be considered as a transformation in a vector space:
In other words, \(v\) is such a vector that \(A\) just changes its scale by a factor \(\lambda\).
In NumPy, eigenvectors and eigenvalues can be obtained by
np.linalg.eigh(<array>) which returns a tuple with the following two
elements:
Eigenvalues (an array of \(\lambda\)) in decreasing order.
Eigenvectors (\(v\)) as a column matrix. I.e.
v[:, i]is the eigenvector corresponding to the eigenvalue \(\lambda[i]\).
Matrix Decompositions
Numpy also provides many frequently used matrix decompositions, as listed below:
Matrix Decomposition |
How to Calculate with NumPy |
|---|---|
Cholesky decomposition |
|
QR factorization |
|
Singular Value Decomposition |
|
Solve a linear system of equations
Given a linear system of equations as follows:
which can be represented in compact form as:
can be solved in NumPy using np.linalg.solve(a, b).
Below is an easy example:
>>> A
array([[0, 1],
[2, 3]])
>>> B
array([3, 4])
>>> np.linalg.solve(A,B)
array([-2.5, 3. ])
>>> X = np.linalg.solve(A,B)
>>> X
array([-2.5, 3. ])
which can be easily verified using multiplication:
>>> np.inner(A, X)
array([3., 4.])
which is equal to B.
10.1.4. Why Use NumPy? Efficiency Benefits¶
The previous sections have illustrated how capable the NumPy library is and how easily we can do many calculations with the pre-defined functionalities e.g. in its linear algebra module. Since we can access each element of an array using Python’s indexing mechanisms, we can be tempted to implementing those functionalities ourselves, from scratch.
However, those functionalities in NumPy are not implemented in Python but in C, another high-level programming language where programs are directly compiled into machine executable binary code. Therefore, they would execute much faster in comparison to what we would be implementing in Python.
The following example illustrates this. When executed, you should see an order of $~$1000 difference in running time.
In other words, you are strongly advised, whenever possible, to use NumPy’s existing functions and routines, and to write every operation in vector or matrix form as much as possible if you are working with matrices.
import numpy as np
from time import time
def matmul_2D(M, N):
"""Custom defined matrix multiplication for two 2D matrices M and N"""
(H_M, W_M) = M.shape
(H_N, W_N) = N.shape
if W_M != H_N:
print("Dimensions of M and N mismatch!")
return None
result = np.zeros((H_M, W_N))
for i in range(H_M):
for j in range(W_N):
for k in range(W_M):
result[i][j] += M[i][k] * N[k][j]
return result
# First let us check that our code works as expected
M = np.random.randn(2,3)
N = np.random.randn(3,4)
print("Our matmul_2D result: \n", matmul_2D(M, N))
print("Correct result: \n", np.matmul(M, N))
# Now let us measure the running-time performances
# Create two 2D large matrices
M = np.random.randn(100, 100)
N = np.random.randn(100, 100)
# Option 1: Use NumPy's matrix multiplication
t1 = time()
result = np.matmul(M, N)
t2 = time()
print("NumPy's matmul took ", t2-t1, "ms.")
# Option 2: Use our matmul_2D function
t1 = time()
result = matmul_2D(M, N)
t2 = time()
print("Our matmul_2D function took ", t2-t1, "ms.")
Our matmul_2D result:
[[-0.24208963 1.18790239 -1.5397112 -1.14521578]
[-2.00697411 4.23675778 -3.74909068 -0.23854542]]
Correct result:
[[-0.24208963 1.18790239 -1.5397112 -1.14521578]
[-2.00697411 4.23675778 -3.74909068 -0.23854542]]
NumPy's matmul took 0.00040411949157714844 ms.
Our matmul_2D function took 1.0593342781066895 ms.
10.2. Scientific Computing with SciPy¶
SciPy is a library that includes many methods and facilities for Scientific Computing. It is closely linked with NumPy so much that NumPy needs to be imported first to be able to use SciPy.
Installation Notes To be able to use the SciPy library, you will need to download it from
scipy.org and install it on your computer. If
you are using a Python package manager (e.g. pip), you can install it
directly using: |
Below is a list of some modules provided by SciPy. Even a brief coverage
of these modules is not feasible in a chapter. However, we will see an
example in Chapter 12 using SciPy’s stats and optimize modules.
Module |
Description |
|---|---|
cluster |
Clustering algorithms |
constants |
Physical and mathematical constants |
fftpack |
Fast Fourier Transform routines |
integrate |
Integration and ordinary differential equation solvers |
interpolate |
Interpolation and smoothing splines |
io |
Input and Output |
linalg |
Linear algebra |
ndimage |
N-dimensional image processing |
odr |
Orthogonal distance regression |
optimize |
Optimization and root-finding routines |
signal |
Signal processing |
sparse |
Sparse matrices and associated routines |
spatial |
Spatial data structures and algorithms |
special |
Special functions |
stats |
Statistical distributions and functions |
10.3. Data handling & analysis with Pandas¶
Pandas is a very handy library for working with files with different formats and analyzing data in different format. To keep things simple, in this section we will just look at CSV files. Note that the facilities for other formats are very similar.
Installation Notes To be able to use the Pandas library, you will need to download it from
pandas.pydata.org and install it on
your computer. If you are using a Python package manager (e.g. pip), you
can install it directly using: |
10.3.1. Supported File Formats¶
As we have seen before, Python already provides facilities for reading and writing files. However, if you need to work with “structured” files such as CSV, XML, XLSX, JSON, HTML, the native facilities of Python would need to extended significantly.
That’s where Pandas comes in. It complements Python’s native facilities to be able to work with structured files for both reading data from them and creating new files. Below is a list of the different file formats that Pandas supports and the corresponding functions for reading or writing them.
Table 10.1. The file formats supported by the Pandas library. The table is adapted from and the reader is encouraged to check the following page to see a more up-to-date list: Pandas IO Reference.
Format Type |
Data Description |
Reader |
Writer |
|---|---|---|---|
text |
CSV |
read_csv |
to_csv |
text |
Fixed-Width Text File |
read_fwf |
|
text |
JSON |
read_json |
to_json |
text |
HTML |
read_html |
to_html |
text |
Local clipboard |
read_clipboard |
to_clipboard |
MS Excel |
read_excel |
to_excel |
|
binary |
OpenDocument |
read_excel |
|
binary |
HDF5 Format |
read_hdf |
to_hdf |
binary |
Feather Format |
read_feather |
to_feather |
binary |
Parquet Format |
read_parquet |
to_parquet |
binary |
ORC Format |
read_orc |
|
binary |
Msgpack |
read_msgpack |
to_msgpack |
binary |
Stata |
read_stata |
to_stata |
binary |
SAS |
read_sas |
|
binary |
SPSS |
read_spss |
|
binary |
Python Pickle Format |
read_pickle |
to_pickle |
SQL |
SQL |
read_sql |
to_sql |
SQL |
Google BigQuery |
read_gbq |
to_gbq |
10.3.2. Data Frames¶
Pandas library is built on top of a data type called DataFrame which
is used to store all types of data while working with Pandas. In the
following, we will illustrate how you can get your data into a
DataFrame object:
1. Loading data from a file. If you have your data in a file that is
supported by Pandas, you can use its reader for directly obtaining a
DataFrame object. Let us look at an example CSV file:
# Download an example CSV file:
!wget -nc https://raw.githubusercontent.com/sinankalkan/CENG240/master/figures/ch10_example.csv
# Import the necessary libraries
import pandas as pd
# Read the file named 'ch10_example.csv'
df = pd.read_csv('ch10_example.csv')
# Print the CSV file's contents:
print("The CSV file contains the following:\n", df, "\n")
# Check the types of each column
df.dtypes
--2021-06-21 20:22:24-- https://raw.githubusercontent.com/sinankalkan/CENG240/master/figures/ch10_example.csv
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.109.133, 185.199.110.133, 185.199.108.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.109.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 162 [text/plain]
Saving to: ‘ch10_example.csv’
ch10_example.csv 100%[===================>] 162 --.-KB/s in 0s
2021-06-21 20:22:25 (2.53 MB/s) - ‘ch10_example.csv’ saved [162/162]
The CSV file contains the following:
Name Grade Age
0 Jack 40.2 20
1 Amanda 30.0 25
2 Mary 60.2 19
3 John 85.0 30
4 Susan 70.0 28
5 Bill 58.0 28
6 Jill 90.0 27
7 Tom 90.0 24
8 Jerry 72.0 26
9 George 79.0 22
10 Elaine 82.0 23
Name object
Grade float64
Age int64
dtype: object
Note the following critical details:
read_csvfile automatically understood that our CSV file had a header (“Name”, “Grade” and “Age”). If your file does not have a header, you can callread_csvwith theheaderparameter set toNoneas follows:pd.read_csv(filename, header=None).read_csvread all columns in the CSV file. If you wish to load only some of the columns (e.g. ‘Name’, ‘Age’ in our example), you can relay this using theusecolsparameter as follows:pd.read_csv(filename, usecols=[ 'Name', 'Age).
2. Convert Python data into a ``DataFrame``. Alternatively, you can
have already a Python data object which you can provide as argument to a
DataFrame constructor as illustrated with the following example:
lst = [('Jack', 40.2, 20), ('Amanda', 30, 25), ('Mary', 60.2, 19)]
df = pd.DataFrame(data = lst, columns=['Name', 'Grade', 'Age'])
print(df)
Name Grade Age
0 Jack 40.2 20
1 Amanda 30.0 25
2 Mary 60.2 19
In many cases, we will require the rows to be associated with names, or sometimes called as keys. For example, instead of referring to a row as “the row at index 1”, we might require accessing a row with a non-integer value. This can be achieved as follows (note how the printed DataFrame looks different):
names = ['Jack', 'Amanda', 'Mary']
lst = [(40.2, 20), (30, 25), (60.2, 19)]
df = pd.DataFrame(data = lst, index=names, columns=['Grade', 'Age'])
print(df)
Grade Age
Jack 40.2 20
Amanda 30.0 25
Mary 60.2 19
Alternatively, we can obtain a DataFrame from a dictionary, which might be easier for us to create data column-wise – note that the column names are obtained from the keys of the dictionary:
d = {'Grade': [40.2, 30, 60.2],
'Age': [20, 25, 19]}
names = ['Jack', 'Amanda', 'Mary']
df = pd.DataFrame(data = d, index=names)
print(df)
Grade Age
Jack 40.2 20
Amanda 30.0 25
Mary 60.2 19
10.3.3. Accessing Data with DataFrames¶
We can access data columnwise or row-wise.
1. Columnwise access.
For columnwise access, you can use df["Grade"], which returns a
sequence of values. To access a certain element in that sequence, you
can either use an integer index (as in df["Grade"][1]) or named
index if it has been defined (as in df["Grade"]["Amanda"]). The
following example illustrates this for the example DataFrame we have
created above:
>>> print(df)
Grade Age
Jack 40.2 20
Amanda 30.0 25
Mary 60.2 19
>>> print(df['Grade'][1])
30.0
>>> print(df['Grade']['Amanda'])
30.0
2. Rowwise access.
To access a particular row, you can either use integer indexes with
df.iloc[<row index>] or df.loc[<row name>] if a named index is
defined. This is illustrated below:
>>> print(df)
Grade Age
Jack 40.2 20
Amanda 30.0 25
Mary 60.2 19
>>> print(df.iloc[1])
Grade 30.0
Age 25.0
Name: Amanda, dtype: float64
>>> print(df.loc['Amanda'])
Grade 30.0
Age 25.0
Name: Amanda, dtype: float64
You can also provide both column index and name index in a single
operation, i.e. [<row index>, <column index>] or
[<row name>, <column name>] as illustrated below:
>>> df.loc['Amanda','Grade']
30.0
>>> df.iloc[1, 1]
25
While accessing a DataFrame with integer indexes, you can also use
Python’s slicing; i.e. [start:end:step] indexing.
10.3.4. Modifying Data with DataFrames¶
Although modifying data in a DataFrame is simple, you need to be
careful about one crucial concept: Let us say you use column and row
names to change the contents of a cell and you access first the column
and then the corresponding row as follows:
>>> print(df)
Grade Age
Jack 40.2 20
Amanda 30.0 25
Mary 60.2 19
>>> df['Grade']['Amanda'] = 45
/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:6: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
This is called chained indexing and Pandas will give you a warning
that you are modifying a DataFrame that was returned while
accessing columns or rows in your original DataFrame:
dfGrade returns a direct access for the original Grade
column or a copy to it and depending on how your data is structured and
stored in the memory, the end result may be different and your change
might not be reflected on the original DataFrame.
Instead, you should use a single access operation as illustrated below
(you may do this with loc and iloc as well):
>>> print(df)
Grade Age
Jack 40.2 20
Amanda 30.0 25
Mary 60.2 19
>>> df.loc['Amanda','Grade'] = 45
>>> df.iloc[1,1] = 30
>>> print(df)
Grade Age
Jack 40.2 20
Amanda 45.0 30
Mary 60.2 19
With these facilities, you can now access every item in a DataFrame
and modify them.
10.3.5. Analyzing Data with DataFrames¶
Once we have our data in a DataFrame, we can use Pandas’s built-in
facilities for analyzing our data. One very simple way to analyze data
is via descriptive statistics, which you can access with
df.describe() and df[<column>].value_counts():
print(df)
df.describe()
Grade Age
Jack 40.2 20
Amanda 30.0 25
Mary 60.2 19
| Grade | Age | |
|---|---|---|
| count | 3.000000 | 3.000000 |
| mean | 43.466667 | 21.333333 |
| std | 15.362725 | 3.214550 |
| min | 30.000000 | 19.000000 |
| 25% | 35.100000 | 19.500000 |
| 50% | 40.200000 | 20.000000 |
| 75% | 50.200000 | 22.500000 |
| max | 60.200000 | 25.000000 |
Apart from these descriptive functions, Pandas provides functions for
sorting (using .sort_values() function), finding the maximum or the
minimum (using .max() or .min() functions) or finding the
largest or smallest n values (using .nsmallest() or .nlargest()
functions:
print("Maximum grade is: ", df['Grade'].max())
print("\nRecords sorted according to age:\n", df.sort_values(by="Age"))
print("\n\nTop two grades are:\n", df['Grade'].nlargest(2))
Maximum grade is: 60.2
Records sorted according to age:
Grade Age
Mary 60.2 19
Jack 40.2 20
Amanda 30.0 25
Top two grades are:
Mary 60.2
Jack 40.2
Name: Grade, dtype: float64
Note that, the displayed output includes the names because we selected names as the indices for accessing the rows.
10.3.6. Presenting Data in DataFrames¶
Pandas provides a very easy mechanism for plotting your data via
.plot() function. Examples are provided below to illustrate how you
can plot your own data. This plotting facility is provided by the
Matplotlib which we will see next. If you are not using Colab, you need
to use the following method to show the plot:
df.plot().figure.show()
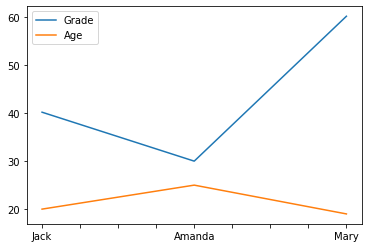
#Plots all columns in different colours
df.plot()
<AxesSubplot:>
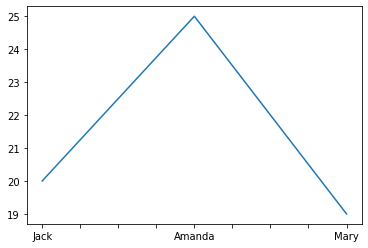
# Plots a single column
df['Age'].plot()
<AxesSubplot:>
10.4. Plotting data with Matplotlib¶
Matplotlib is a very capable library for drawing different types of plots in Python. It is very well integrated with Numpy, Scipy and Pandas, and therefore, all these libraries are very frequently used together seamlessly.
Installation Notes To be able to use the matplotlib library, you will need to download it
from matplotlib.org and install it on your
computer. If you are using a Python package manager (e.g. pip), you can
install it directly using: |
10.4.1. Parts of a Figure¶
A figure consists of the following elements (see also Fig. 10.4.1): - Title of the figure. - Axes, together with their ticks, tick labels, and axis labels. - The canvas of plot, which consists of a drawing of your data in the form of a dots (scatter plot), lines (line plot), bars (bar plot), surfaces etc. - Legend, which informs the perceiver about the different plots in the canvas.
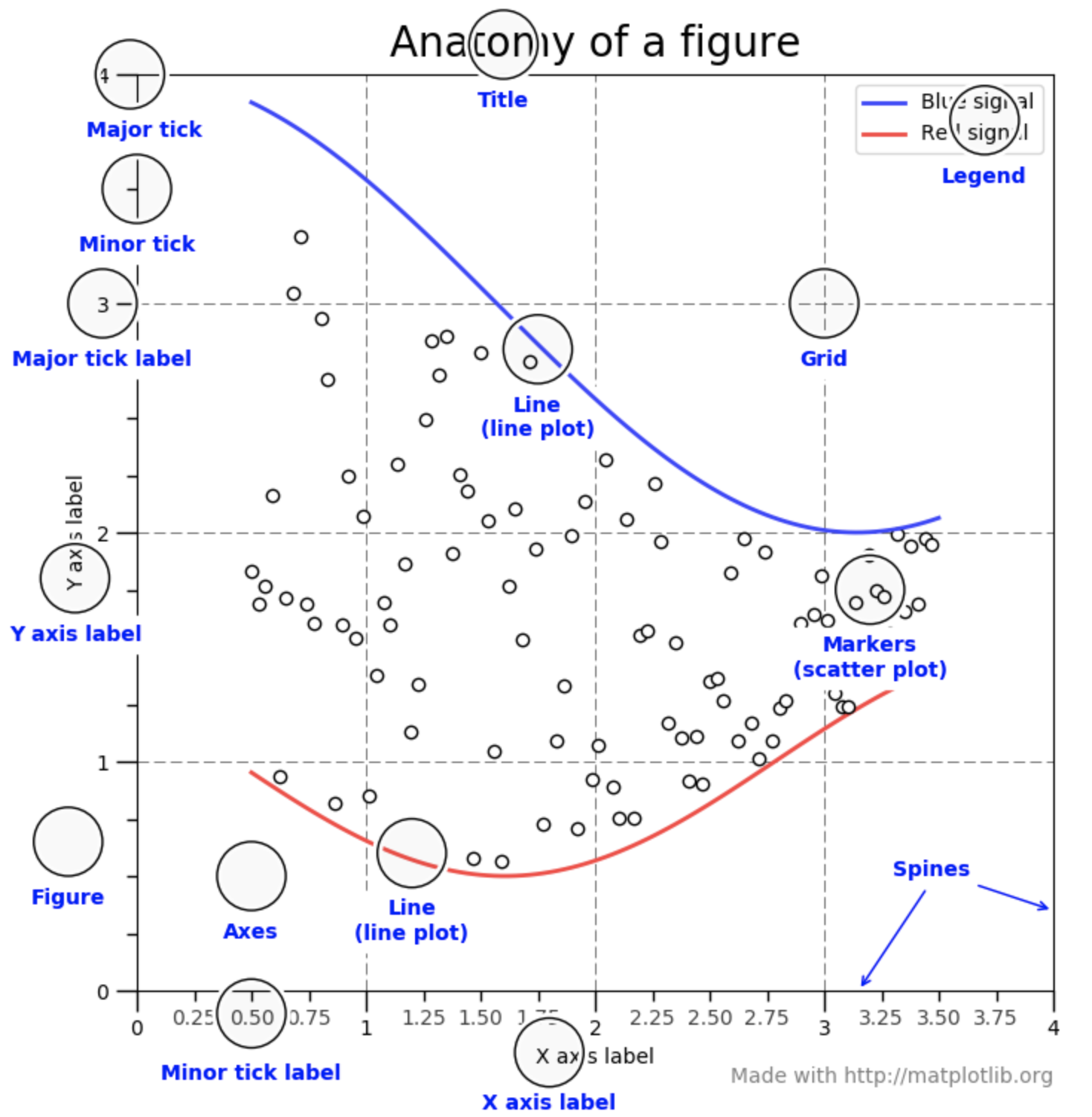
Fig. 10.4.1 A figure consists of several components all of which you can change in matplotlib easily. Figure source: Matplotlib Usage Guides.¶
10.4.2. Preparing your Data for Plotting¶
Matplotlib expects numpy arrays as input and therefore, if you have your
data in a numpy array, you can directly plot it without any data type
conversion. With pandas DataFrame, the behavior is not guaranteed
and therefore, it is recommended that the values in a DataFrame are
first converted to a numpy array, using e.g.:
print(df)
age_array = df['Age'].values
print("The `Age` values in an array form are:", age_array)
print("The type of our new data is: ", type(age_array))
Grade Age Jack 40.2 20 Amanda 30.0 25 Mary 60.2 19 The Age values in an array form are: [20 25 19] The type of our new data is: <class 'numpy.ndarray'>
10.4.3. Drawing Single Plots¶
There are two ways to plot with matplotlib: In an object-oriented style or the so-called Pyplot style:
1. Drawing in an Object-Oriented Style.
In this style, we create a figure object and an axes object and work with those to create our plots. This is illustrated in the following example:
import matplotlib.pyplot as plt
import numpy as np
# Uniformly sample 50 x values between -2 and 2:
x = np.linspace(-2, 2, 50)
# Create an empty figure
fig, ax = plt.subplots()
# Plot y = x
ax.plot(x, x, label='$y=x$')
# Plot y = x^2
ax.plot(x, x**2, label='$y=x^2$')
# Plot y = x^3
ax.plot(x, x**3, label='$y=x^3$')
# Set the labels for x and y axes:
ax.set_xlabel('x')
ax.set_ylabel('y')
# Set the title of the figure
ax.set_title("Our First Plot -- Object-Oriented Style")
# Create a legend
ax.legend()
# Show the plot
# fig.show() # Uncomment if not using Colab
<matplotlib.legend.Legend at 0x125532250>
2. Drawing in a Pyplot Style. In the Pyplot style, we directly call
functions in the pyplot module (matplotlib.pyplot) to create a
figure and draw our plots. This style does not work with an explicit
figure or axes objects. This is illustrated in the following example:
# Uniformly sample 50 x values between -2 and 2:
x = np.linspace(-2, 2, 50)
# Plot y = x
plt.plot(x, x, label='$y=x$')
# Plot y = x^2
plt.plot(x, x**2, label='$y=x^2$')
# Plot y = x^3
plt.plot(x, x**3, label='$y=x^3$')
# Set the labels for x and y axes:
plt.xlabel('x')
plt.ylabel('y')
# Set the title of the figure
plt.title("Our First Plot -- Pyplot Style")
# Create a legend
plt.legend()
# Show the plot
#plt.show() # Uncomment if not using Colab
<matplotlib.legend.Legend at 0x1256ab9a0>
10.4.4. Drawing Multiple Plots in a Figure¶
In many situations, you will need to draw multiple plots side by side in
a single figure. This can be performed in the object-oriented style
using the subplots() function to create a grid and then use the
created subplots and axes to draw the plots. This is illustrated with an
example below:
# Create a 2x2 grid of plots
fig, axes = plt.subplots(2, 2)
# Plot (1,1)
axes[0,0].plot(x, x)
axes[0,0].set_title("$y=x$")
# Plot (1,2)
axes[0,1].plot(x, x**2)
axes[0,1].set_title("$y=x^2$")
# Plot (2,1)
axes[1,0].plot(x, x**3)
axes[1,0].set_title("$y=x^3$")
# Plot (2,2)
axes[1,1].plot(x, x**4)
axes[1,1].set_title("$y=x^4$")
# Adjust vertical space between rows
plt.subplots_adjust(hspace=0.5)
# Show the plot
#fig.show() # Uncomment if not using Colab
It is possible to do this in the PyPlot style as well, as illustrated below:
# Plot (1,1)
plt.subplot(2, 2, 1)
plt.plot(x, x)
plt.title('$y=x$')
# Plot (1,2)
plt.subplot(2, 2, 2)
plt.plot(x, x**2)
plt.title('$y=x^2$')
# Plot (2,1)
plt.subplot(2, 2, 3)
plt.plot(x, x**3)
plt.title('$y=x^3$')
# Plot (2,2)
plt.subplot(2, 2, 4)
plt.plot(x, x**4)
plt.title('$y=x^4$')
# Adjust vertical space between rows
plt.subplots_adjust(hspace=0.5)
# Show the plot
#plt.show() # Uncomment if not using Colab
10.4.5. Changing elements of a plot¶
All elements that are visualized in Figure 10.4.1 can be changed in
matplotlib. We will skip these details to keep our focus in the book to
the practical uses of these libraries. The interested reader can look up
the extensive documentation at
https://matplotlib.org/2.1.1/contents.html or look at the help page of a
function (e.g. help(plt.plot)) to see how to modify all elements of
a figure.
10.5. Important Concepts¶
We would like our readers to have grasped the following crucial concepts and keywords from this chapter:
NumPy arrays and their properties: array shape, dimensions, sizes, elements.
Accessing and modifying elements of a NumPy array.
Simple algebraic functions on NumPy arrays.
SciPy and its basic capabilities.
Pandas, DataFrame, loading files with Pandas.
Accessing and modifying content in DataFrames.
Analyzing and presenting data in DataFrames.
Matplotlib and different ways to make plots.
Drawing single and multiple plots. Changing elements of a plot.
10.6. Further Reading¶
NumPy documentation: https://numpy.org/doc/
SciPy documenation: https://docs.scipy.org/doc/scipy/reference/
Pandas documentation: https://pandas.pydata.org/docs/
Matplotlib documentation: https://matplotlib.org/2.1.1/contents.html
Introduction to Algebra with Python: https://pabloinsente.github.io/intro-linear-algebra
10.7. Exercises¶
Define functions that work like the
sum,mean,minandmaxoperations provided by NumPy. These functions should take a single 2D array and return the result as a number. You can assume that the operation applies to the whole array and not to a single axis.Create a simple CSV file using your favorite spreadsheet editor (e.g. Microsoft Excel or Google Spreadsheets) and create a file with your exams and their grades as two separate columns. Save the file, upload it to the Colab notebook and do the following:
Load the file using Pandas.
Calculate the mean of your exam grades.
Calculate the standard deviation of your grades.
Using Matplotlib, generate the following plots with suitable names for the axes and the titles.
Draw the following four functions in separate single plots: \(\sin(x), \cos(x), \tan(x), \cot(x)\).
Draw these four functions in a single plot.
Draw a multiple 2x2 plot where each subplot is one of the four functions.
